About scDrugMap
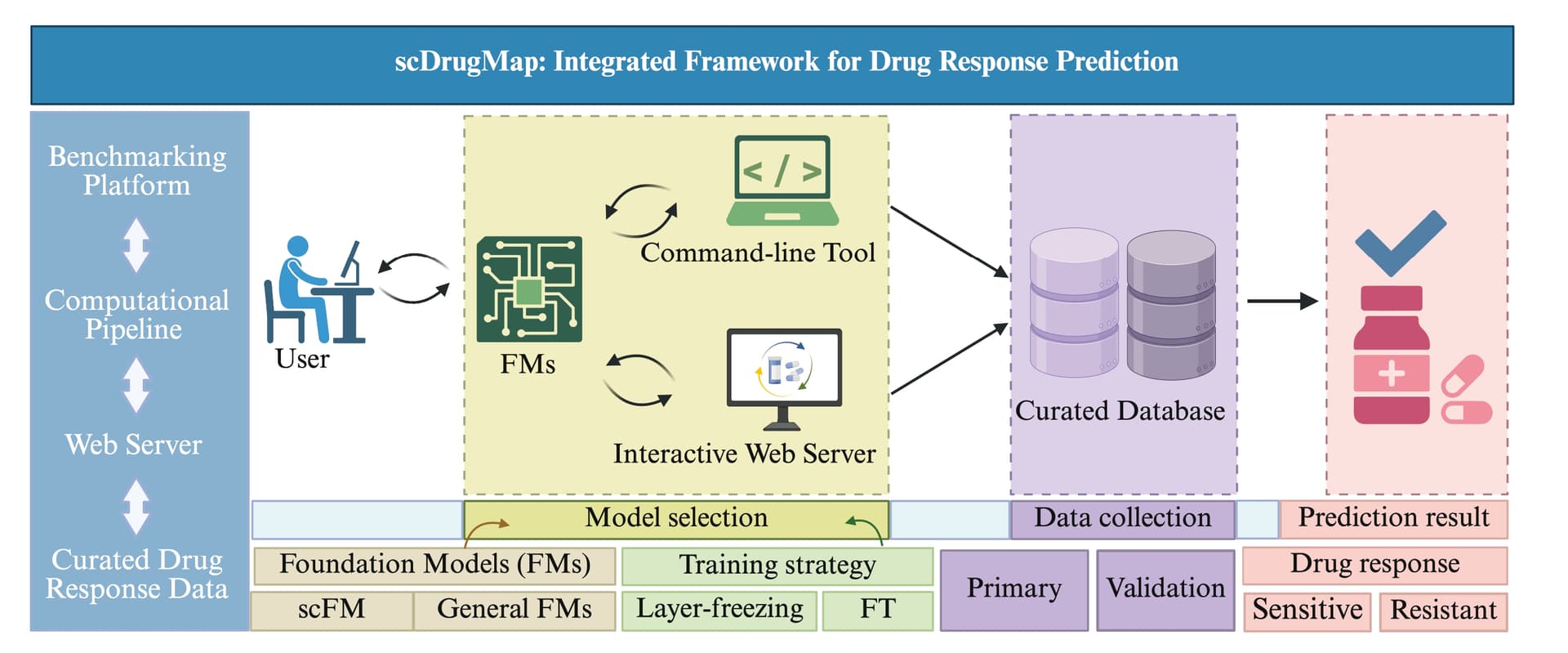
scDrugMap is a scalable and user-friendly framework for predicting drug responses in single-cell transcriptomics data using large-scale foundation models. It supports both a Python command-line tool and an interactive web interface (scdrugmap.com) to facilitate drug discovery and translational research.
Overview
Drug resistance remains a major challenge in cancer treatment. Single-cell profiling provides critical insights into cellular heterogeneity and drug sensitivity, but high dimensionality and data sparsity can complicate downstream analysis.
scDrugMap evaluates and integrates the predictive power of eight single-cell foundation models and two natural language models across diverse cancer types and treatment regimens. It supports:
- Pooled-data and cross-data model evaluation
- Layer freezing and LoRA fine-tuning
- Zero-shot and fine-tuned inference
- Single-cell resolution predictions across 326,000+ cells and 36+ datasets
Specific Instructions
1. Select LLM Model
Choose the large language model (LLM) you want to use for drug response prediction from the dropdown menu.

2. Upload CSV/TSV File
Upload a tab-delimited gene expression matrix (.tsv, .csv, .txt) for single cells:
- Each row = one single cell
- Cell_barcode: Unique cell ID (e.g., C70R_C70R.bcDWVD)
- Condition: Experimental condition (e.g., resistant, sensitive)
- Remaining columns: Gene symbols (e.g., AAAS, AACS, …) with integer counts
The example input file can be downloaded and used for testing purpose.

3. Upload & Preprocess Data
After selecting your file, click the Upload & Preprocess Data button. The system will automatically preprocess the data for analysis.

4. Choose Prediction Mode
You can either:
- Fixed embeddings: Fast prediction of accuracy, precision, and F1 score.
- Fine-tuning: Train the model on your uploaded data for better performance (slower).
Adjustable parameters:
- Number of Epochs
- Training Rate
- Learning Rate

The training progress, loss curves, and metrics are displayed in real time.

5. Download Trained Model
Once training is complete, click Download Model to save the fine-tuned model to your local machine, also Download Test Records to save the test records to your local machine.
